CEDAR Release 2.4
We released version 2.4 of the CEDAR Workbench on September 6, providing more user features and enhancements.
OpenView offers public option for CEDAR artifacts
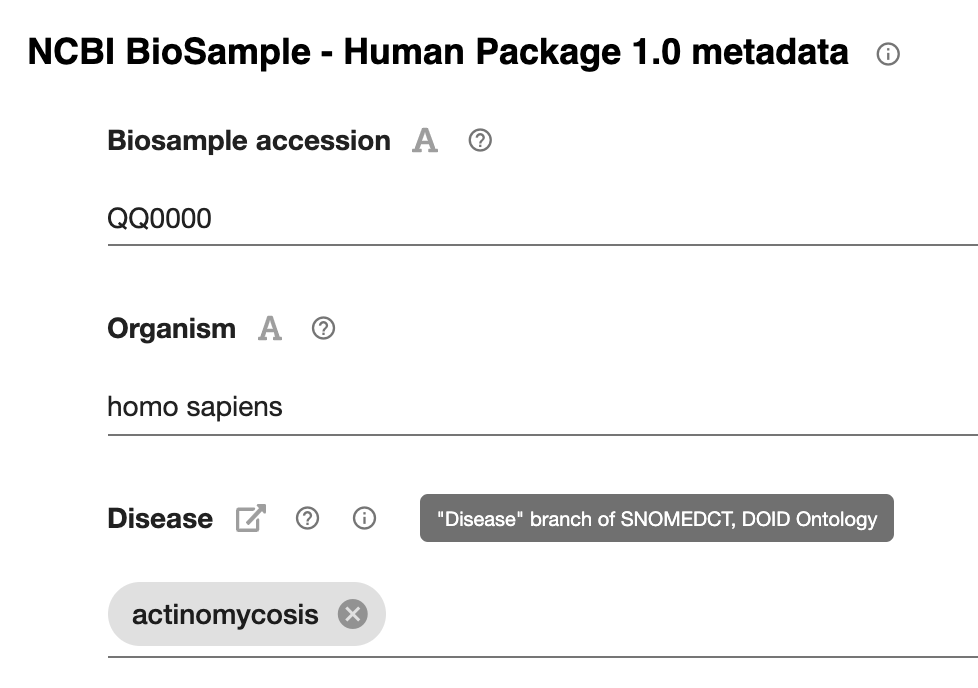 Did you ever want to show your template or metadata values to a colleague, without logging in? Do you want to view all your metadata on the web? Or maybe you’d like an IRI that anyone can use to see your work?
Did you ever want to show your template or metadata values to a colleague, without logging in? Do you want to view all your metadata on the web? Or maybe you’d like an IRI that anyone can use to see your work?
Now you can make your CEDAR artifact—metadata instance, template, element, or field—visible on the web. CEDAR’s OpenView service presents the CEDAR artifact as a publicly visible web page, with pop-up metadata descriptions and access to JSON and RDF views of the content. To make public your template, element, or field, simply enable OpenView from the workspace menu for the artifact. For now, if you want to make your metadata public, the template it’s based on must also be public—we can help you with this.
Instructions for CEDAR’s OpenView feature may be found at its CEDAR manual page.
Find field names in templates, elements, and fields
Adding to CEDAR’s ability to search for field names in CEDAR instances, you can now search for field names in CEDAR templates, elements, and fields.
Just like searching for field names in metadata instances, a colon after the string indicates a search within field names. The syntax ‘namestring:’ in CEDAR’s search bar will find the templates containing ‘namestring’ in the title. In the CEDAR search syntax, an asterisk matches any string and a question mark matches any character:
title:*, or simplytitle: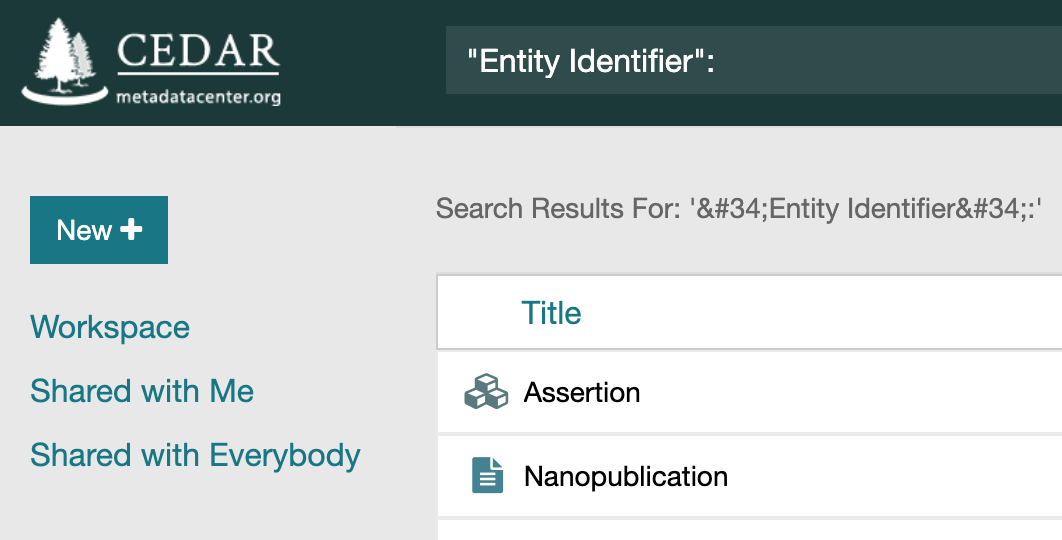
Publish*:"Contact Email":to?ic:
This functionality is documented in more detail here.
AIRR Community NCBI Pipeline
We improved the CAIRR pipeline for submitting MiAIRR data to NCBI. The AIRR community has documented CEDAR-driven MiAIRR submissions to NCBI in the MiAIRR-to-NCBI Submission Manual, the primary user documentation for submitting AIRR metadata. In the SRA section, the pipeline now checks user-entered file names against the names of files actually submitted, and alerts the user if they do not match, and file type options have been updated to reflect NCBI expectations. Finally, members of the AIRR community have validated that submissions appear appropriately in NCBI repositories.
Added Human Tissue NCBI Pipeline
CEDAR added a second NCBI submisssion pipeline for submitting metadata on Human Tissue studies. The template for this pipeline is modeled on the AIRR Community pipeline, but is customized to the NCBI BioSample Human Package 1.0.
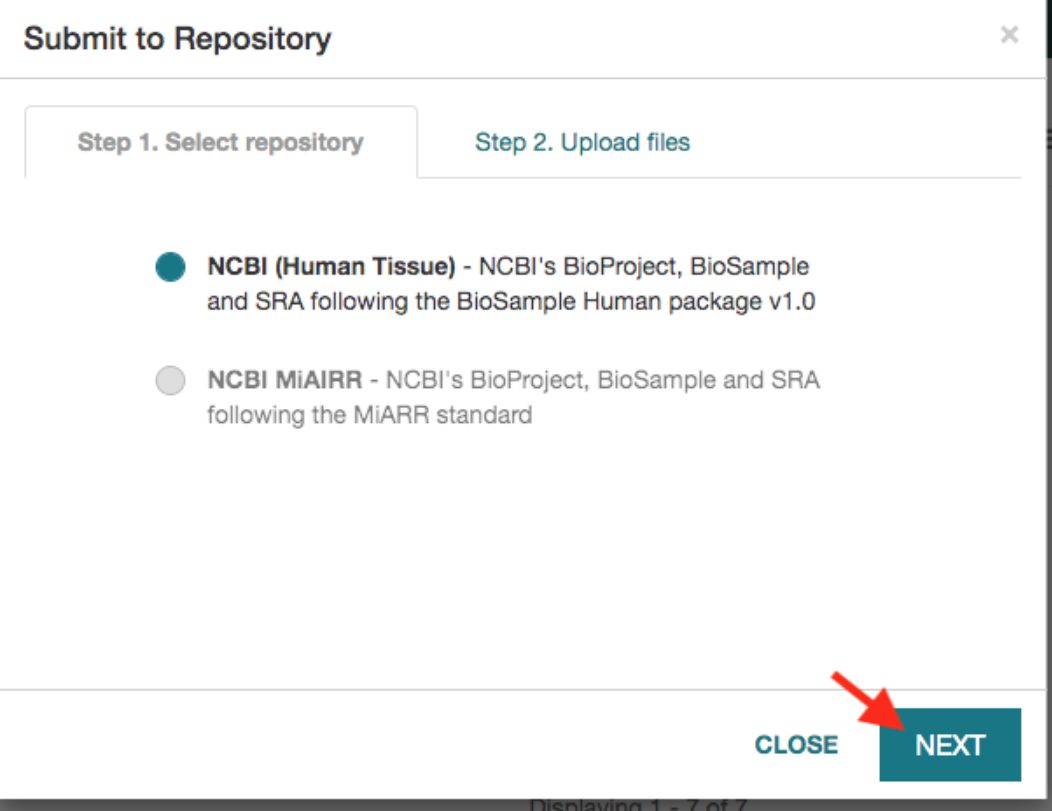 The template and template elements used by this pipeline are publicly available in the following CEDAR folder: All/Shared/Shared by CEDAR/CEDAR-to-NCBI Pipeline. Documentation of the pipeline may be found in the CEDAR pipelines documentation page.
The template and template elements used by this pipeline are publicly available in the following CEDAR folder: All/Shared/Shared by CEDAR/CEDAR-to-NCBI Pipeline. Documentation of the pipeline may be found in the CEDAR pipelines documentation page.
Inside News
Categories for Artifacts
To handle categorization of CDE fields, we have designed a category system that can be used by different communities to categorize CEDAR artifacts according to their own hierarchical labels. The API for this system has been implemented, and its user interface is planned for the next release.
Performance
We improved CEDAR’s performance for more search types in BioPortal class hierarchies.
Inside Inside News
We refactored code to handle resources (artifacts, users, groups, and folders) in a uniform way. And this release also incorporates some bug fixes.
All the tasks completed for the 2.4 release series can be found with this GitHub search, or by visiting the GitHub release page for release-2.4.

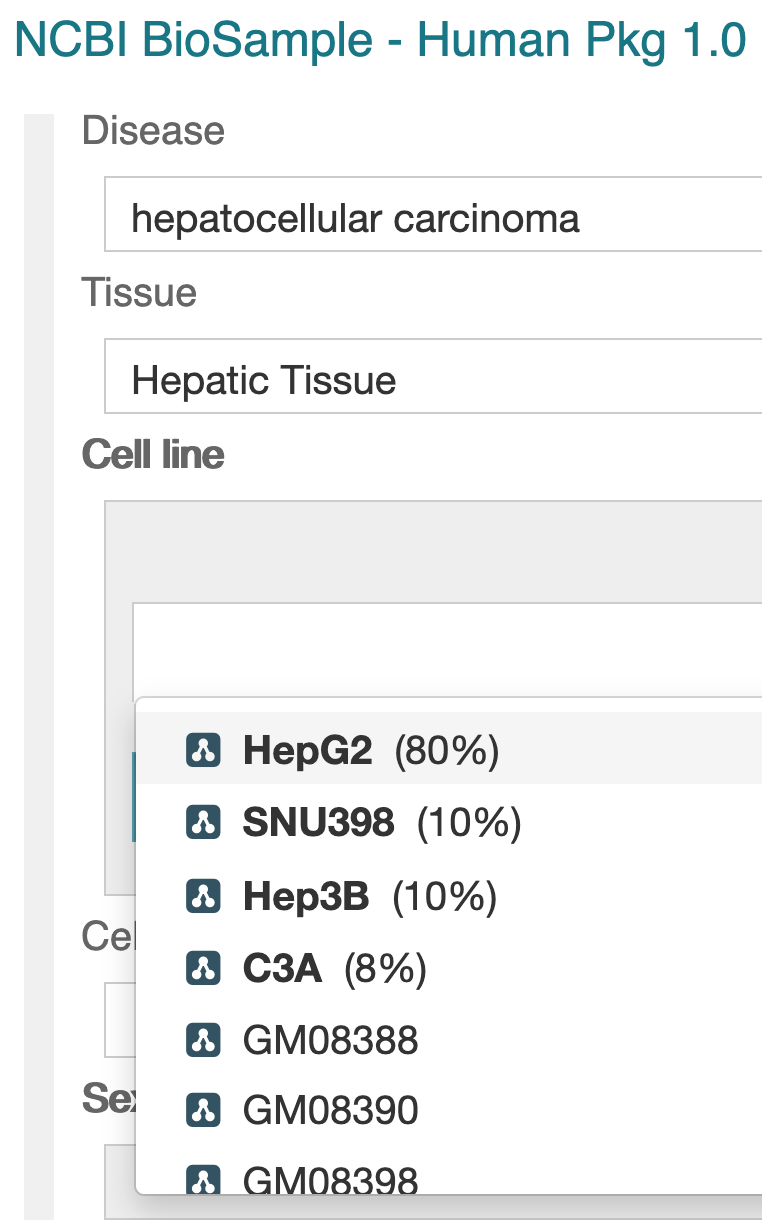 We significantly updated CEDAR’s value recommendation capabilities in this release. The new approach adopts a powerful mechanism to suggest new values for fields in a templates as users fill in those fields. If the template creator enables the value recommender for at least 2 text fields (using each field’s SUGGESTIONS tab), CEDAR will find patterns across all the previously entered values for recommendation-enabled fields. If those patterns are strong enough over many metadata instances, CEDAR will recommend values to a user based on the (enabled) values that the user has already filled in.
We significantly updated CEDAR’s value recommendation capabilities in this release. The new approach adopts a powerful mechanism to suggest new values for fields in a templates as users fill in those fields. If the template creator enables the value recommender for at least 2 text fields (using each field’s SUGGESTIONS tab), CEDAR will find patterns across all the previously entered values for recommendation-enabled fields. If those patterns are strong enough over many metadata instances, CEDAR will recommend values to a user based on the (enabled) values that the user has already filled in.  CEDAR now provides Shared With Me and Shared With Everybody modes on the workspace to let users see globally shared resources, or just resources explicitly shared with them. The default Workspace view just shows the files you own and administer. Shared with Me shows content that someone shared with you directly; and Shared with Everybody shows content that all CEDAR users can see (there are a lot of those!).
CEDAR now provides Shared With Me and Shared With Everybody modes on the workspace to let users see globally shared resources, or just resources explicitly shared with them. The default Workspace view just shows the files you own and administer. Shared with Me shows content that someone shared with you directly; and Shared with Everybody shows content that all CEDAR users can see (there are a lot of those!).