CEDAR Offers Support for CDEs from caDSR
We are pleased to report that CEDAR template creators can now import from over 60,000 of NCI’s caDSR Common Data Elements (CDEs) to build new Fields in CEDAR. Using CEDAR’s search, browsing, and viewing services template builders can easily build a form based partly or entirely on CDEs from caDSR.
Over the last 18 months, CEDAR developers have collaborated with the NCI to adapt CEDAR capabilities to the unique characteristics of CDEs. By representing these CDEs as CEDAR Fields, we have made them fully accessible to CEDAR users. CEDAR already handled many of the specialized features that are found in the caDSR templates, and the CEDAR team added some features to support particular CDE workflows.
User Applications
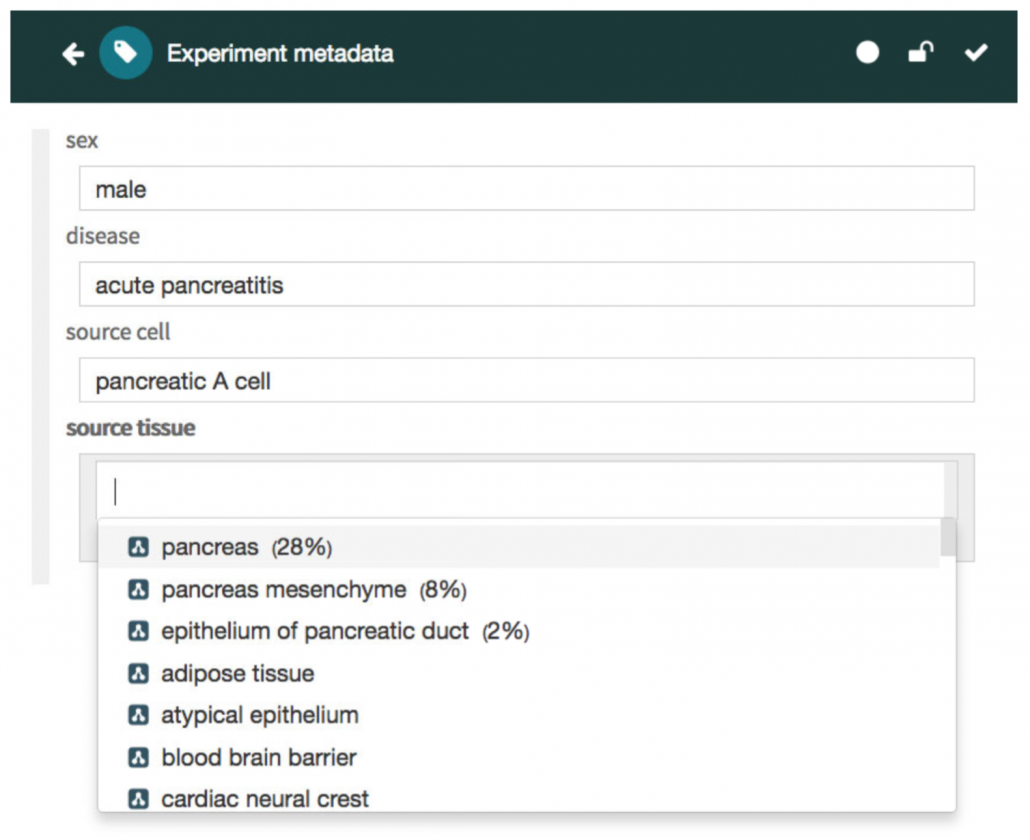
CEDAR’s easy-to-use system for working with metadata templates and reusable components work with the imported CDEs in several ways. A CEDAR Template creator may import CDE representations into a Template using CEDAR’s Template Designer import process, in this way building a metadata form based partly or entirely on CDE content from caDSR.
All the resource discovery and lookup features in CEDAR also work for CDEs. So you can use CEDAR’s search, browsing, and viewing services to find and review CDE content in the system. And CEDAR’s REST APIs also work for the CDEs in CEDAR, which means users can remotely discover and download CDE content.
CEDAR users can even build new Fields by making a copy of any of the CDE-based Fields that are already in CEDAR. The user can modify this copy—which at this point is a generic CEDAR Field—however he or she wants, and will inherit any other Field values like the label, description and help tip. (Eventually CEDAR might support re-submission of CDEs to a CDE repository, but this is not offered at this time.)
About Common Data Elements
CDEs offer precise specifications of questions, including the set of allowable answers to each question. Generally following ISO 11179 data standards, CDEs are decribed in great detail, including information about their development history. CDEs are increasingly being adopted to help improve standardization and interperability, but while CDEs can provide a strong conceptual foundation for interoperation, there are no widely recognized serialization or interchange formats to describe and exchange their definitions.
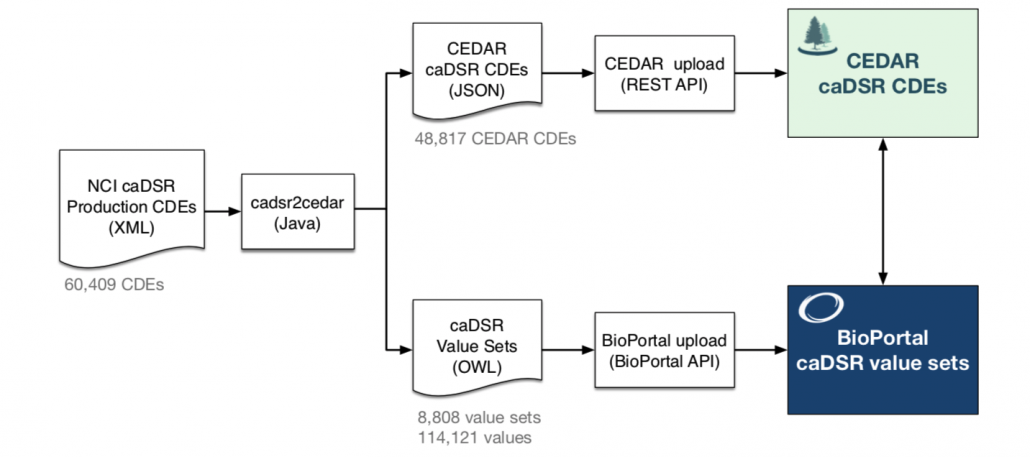
CDE registries can help standardize the way CDEs are collected, stored, transferred, and reported. One of the largest CDE registries has been developed by the U.S. National Cancer Institute (NCI) with the goal of facilitating multidisciplinary, multi-institutional cancer research. This registry is called the Cancer Data Standards Repository (caDSR) and it contains over 60,000 CDEs that cover many aspects of cancer research. The U.S. National Institutes of Health (NIH) are also developing a multi-discipline registry that aims to unify the range of biomedical CDEs that have been produced by a variety of NIH and other organizations (https://cde.nlm.nih.gov).
How CEDAR Adopts CDEs from caDSR
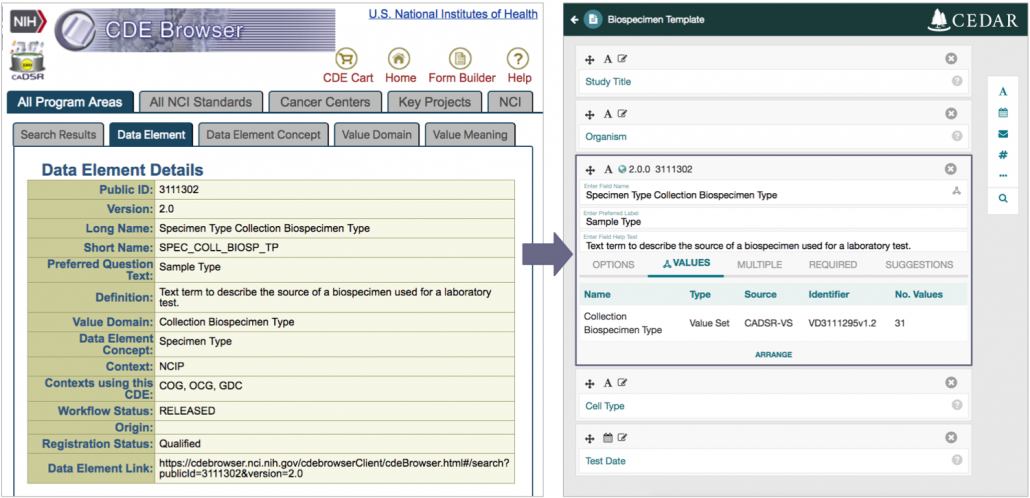
To make existing CDEs more readily accessible to form builders, we extended our CEDAR Web-based metadata management platform to provide a core representation of CDEs suitable for specifying questions in a metadata acquisition system. We do not manage the entire CDE specification—that contains a comprehensive implementation of the ISO/IEC 11179 standard—but focus instead on core functionality that specifies the questions and the values used to answer those questions.
By importing the XML-defined CDEs from the caDSR system into JSON Schema-defined fields in CEDAR, we made these specifications available to any CEDAR user. We run the conversion process automatically to keep the CEDAR CDEs up-to-date with respect to the source content.
Additional Information
You can find more information about CEDAR and its use of CDEs in the following resources.

 In earlier workshops to work on funder metadata, the CEDAR team helped funders describe a
In earlier workshops to work on funder metadata, the CEDAR team helped funders describe a