We believe ease of use is paramount. Because the ability to share data is so important, we want the process of metadata creation and use to be as painless as possible. Our strategic approach addresses the challenges we have seen in other similar projects.
Six key elements contribute to what we believe will be a successful system:
- Interfaces and tools built and tested specifically for metadata creation
- Consistency in terminology
- Machine learning
- Editing capabilities
- Training and outreach
- Building on past work and leveraging ongoing collaborations
Interface and Tools Built and Tested Specifically for Metadata Creation
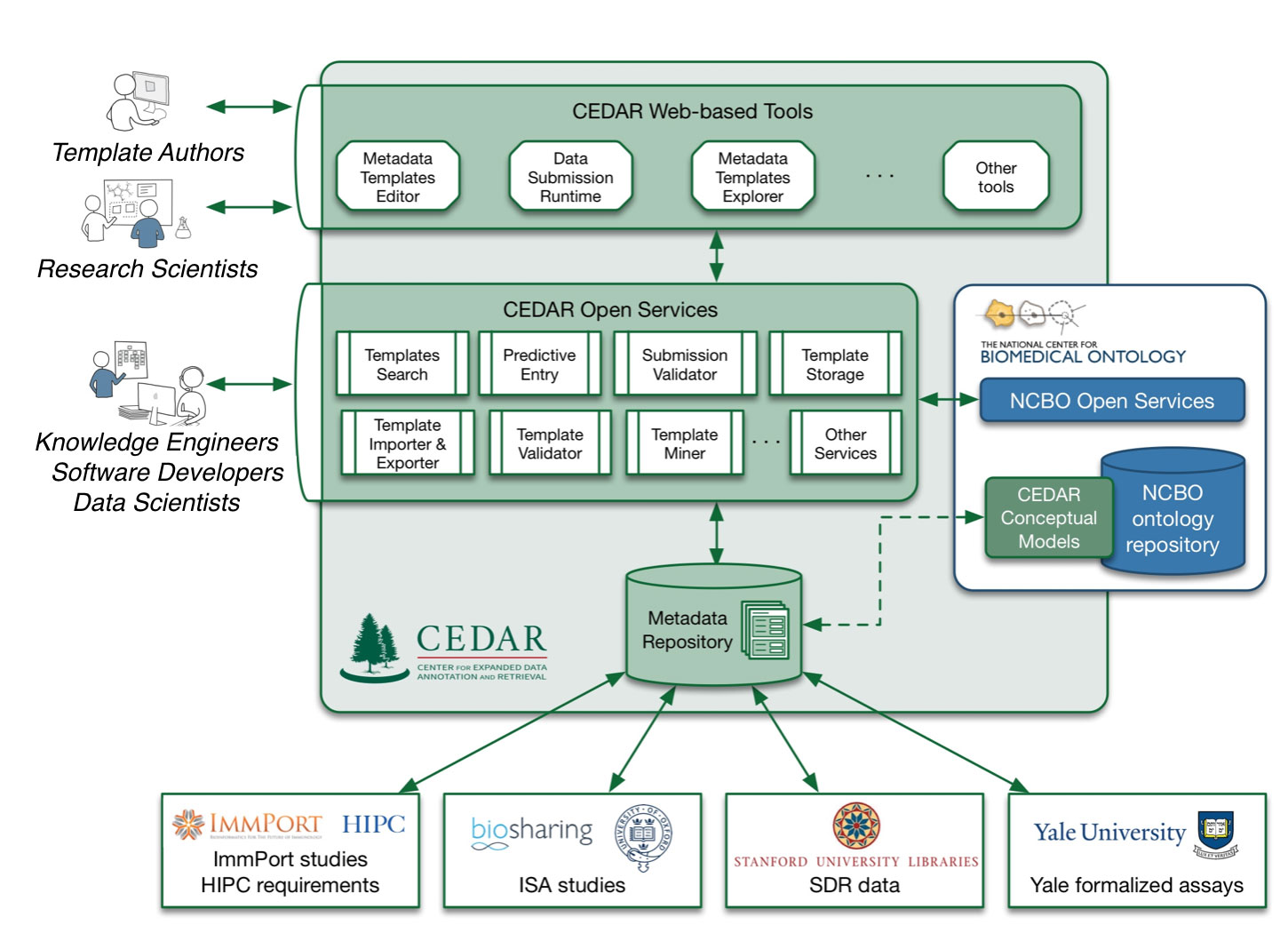 We plan to build a framework that will support not just creating a robust description of an entity, but also creating a corresponding template that can constrain the creation of that description. The template creation, and the instantiated data description that it will produce, will both follow the same principles and strategies, including the ease of use emphasized above.
We plan to build a framework that will support not just creating a robust description of an entity, but also creating a corresponding template that can constrain the creation of that description. The template creation, and the instantiated data description that it will produce, will both follow the same principles and strategies, including the ease of use emphasized above.
Certain types of metadata are shared across disciplines; these include, but are not limited to, names of authors, titles of papers, and language. However, individual disciplines and subspecialties will require inclusion of specific sets of metadata.
Our approach is to create a framework that can support both general and discipline-specific metadata. We envision allowing users to augment the basic set of metadata with other types that address their needs. As the number of available datasets grows, the system will be able to suggest appropriate categories of metadata to include based on the user’s discipline. This will streamline the process of metadata creation.
Consistency in Terminology
An important feature of the system will be the development of controlled knowledge resources—ontologies, value sets, vocabularies, data concept elements (DCEs), and others—that are appropriate for different disciplines and types of research. We will build on existing ontologies, such as the Unified Medical Language System, and also enable users to discover, create, and manage their own knowledge resources.
A controlled set of semantic options eases the process of metadata entry, as the system provides relevant choices to users based on discipline and subject matter, and suggests the best choices based on ‘intelligent’ features like auto-completion, predictive correlations, and user history. Further, users can see more information about their choices, and make better selections based on the context that can be provided.
The consistent use of controlled terminology aids researchers searching for data as well. Not only will searches be able to find the common terms that have been suggested as the metadata is created, those terms can be related to other concepts using semantic technologies, and searches can leverage those relationships to identify other related materials.
Machine Learning
We will use machine-learning techniques to facilitate both predictive entry of metadata and metadata quality assurance. By analyzing past entries, we can develop predictions of not just the likeliest values for given fields, but the fields that are most likely to be needed for a particular data set, and even for a particular template.
Through a multi-pronged approach to integration of our analytic results, we will work to make it simple, educational, and maybe even fun, for scientists to annotate their experimental data in ways that will ensure their value to the scientific community.
Editing Capabilities
 The ability to edit metadata after its initial creation is essential. New discoveries will inevitably uncover important links between datasets that previously seemed unrelated. Terminology will naturally change over time, as knowledge and language evolve. Individual datasets and studies do not exist in isolation, but become related to other artifacts through citation and commentary.
The ability to edit metadata after its initial creation is essential. New discoveries will inevitably uncover important links between datasets that previously seemed unrelated. Terminology will naturally change over time, as knowledge and language evolve. Individual datasets and studies do not exist in isolation, but become related to other artifacts through citation and commentary.
Editing will allow researchers to show relationships between different datasets and papers. Examples include:
- Meta-analyses of multiple datasets or papers;
- Letters to the editor and responses to a particular paper;
- Further research that reproduces or builds on results from a previous study; and
- Semantic analyses discovering common entities across multiple domains, entities, or even entity types (e.g., database entries, papers, and ontologies).
Training and Outreach
To ensure acceptance and success of the new framework, we plan to train the community in the use of metadata and our system. Our training mission will be broad, including among other activities:
- The education of graduate students and post-doctoral fellows in the use of community standards and semantic technology in support of Big Data research;
- The instruction of biomedical investigators in the use of our Center’s technology;
- The instruction of investigators in the use of metadata technologies generally; and
- The indoctrination of the research community in the importance of metadata and the benefits that high-quality data annotation offer the scientific process.
Building on Past Work and Leveraging Ongoing Collaborations
We do not seek to “reinvent the wheel”—CEDAR builds on several longstanding projects of national scope that provide critical ideas, existing standards, related services, and robust software.
As a part of NIH’s Big Data to Knowledge (BD2K) initiative, the CEDAR center will leverage the community of investigators working on common interests and solutions. This community will provide both technical expertise and solutions, and user communities to adopt and provide feedback on our own solutions.
Our work takes advantage of the NIH’s 10‑year investment in the National Center for Biomedical Ontology (NCBO) under its program of National Centers for Biomedical Computing, and builds on the NIH’s 25‑year investment in the Protégé resource at Stanford University.
Our long‑standing collaboration with the BioSharing and ISA efforts—and their involvement with the new data publication platform promoted by the Nature Publishing Group—provides additional support.
We will validate our work in the context of ImmPort, the NIAID’s repository of all datasets developed from grants in its portfolio, offering us a comprehensive and vibrant microcosm in which to test our ideas and to learn from dedicated collaborators.

